Dr. Oki’s paper has been published in Epigenetics & Chromatin.
Elucidating disease-associated mechanisms triggered by pollutants via the epigenetic landscape using large-scale ChIP-Seq data
Zhaonan Zou, Yuka Yoshimura, Yoshihiro Yamanishi, Shinya Oki
Epigenetics & Chromatin 2023 Sep 25;16:34
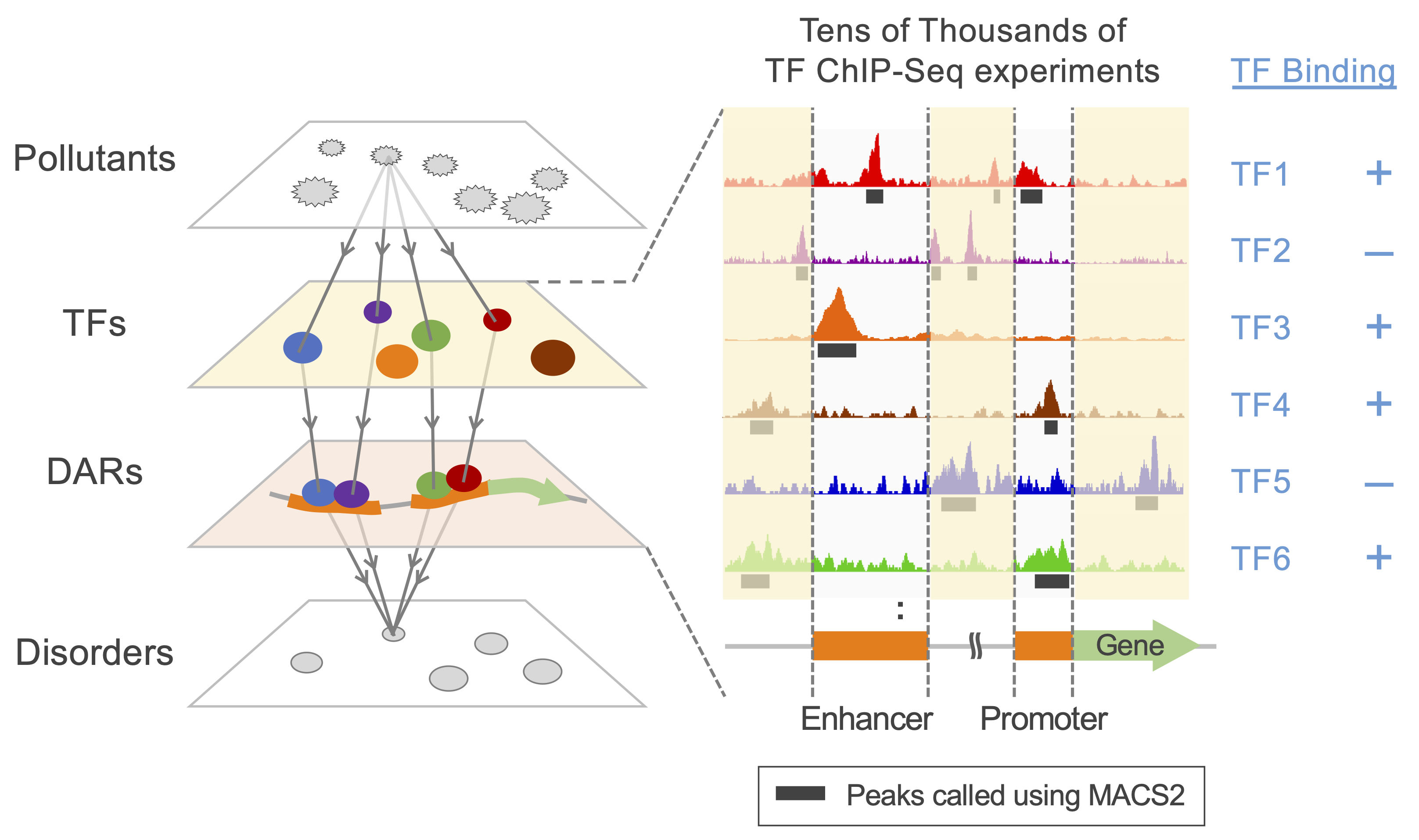
Despite well-documented effects on human health, the action modes of environmental pollutants are incompletely understood. Transcriptome-based approaches are widely used to predict associations between chemicals and disorders. However, the molecular cues regulating gene expression remain unclear. To elucidate the action modes of pollutants, we proposed a data-mining approach combining epigenome (ATAC-Seq) and large-scale public ChIP-Seq data from ChIP-Atlas to identify transcription factors that are enriched in pollutant-induced differentially accessible genomic regions (DARs), thereby integratively regulating gene expression upon pollutant exposure. By using the proposed approach, we predicted that PM2.5 inhibit the binding of hematopoietic differentiation regulator to the genome, thereby perturbing normal blood cell differentiation and leading to immune dysfunction; and lead induces fatty liver by disrupting the normal regulation of lipid metabolism by altering hepatic circadian rhythms. Thus, our approach has the potential to reveal pivotal TFs that mediate adverse effects of pollutants, thereby facilitating the development of strategies to mitigate environmental pollution damage.