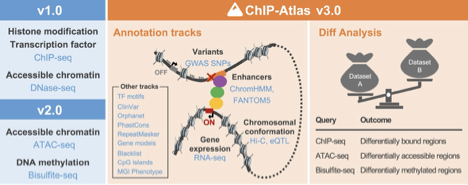
ChIP-Atlas 3.0: a data-mining suite to explore chromosome architecture together with large-scale regulome data
Zhaonan Zou, Tazro Ohta, Shinya Oki
Nucleic Acids Research 2024 May 16;gkae358
The regulation of gene expression in eukaryotes is precisely orchestrated in a spatiotemporal manner by cell-specific epigenomic states, genome-protein interactions, and histone modification patterns. To elucidate these regulatory mechanisms, we have been developing ChIP-Atlas (https://chip-atlas.org), an integrative epigenomic database, for several years. This database visualizes genome-protein interactions (ChIP-seq), chromatin accessibility (ATAC-seq), and methylome information (Bisulfite-seq) for humans and five model organisms (mouse, rat, fruit fly, nematode, and budding yeast), and provides data-mining tools to fully utilise these data. A major update has recently been implemented in ChIP-Atlas, which now incorporates annotation tracks such as three-dimensional chromosomal architectures (Hi-C) and disease-associated genomic variants (GWAS SNPs). Furthermore, the Diff Analysis tool has been introduced for comparative analyses on epigenomic data sets. Consequently, ChIP-Atlas has developed into an epigenomic information infrastructure, comprising the world’s largest data set (376,000 experiments). This is expected to contribute further to the elucidation of the mechanisms of hereditary disease onset, modes of drug action, and the efficiency of studies on cellular differentiation and direct reprogramming.