We will reveal mechanisms for the long-term maintenance of chromosome functions and their aging-associated deterioration by using the mouse as a model, with pioneering efforts to establish the naked mole rat as a new model for germ cell research.
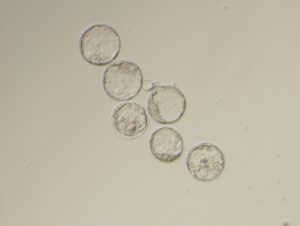
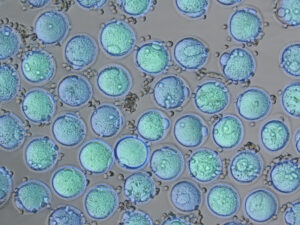
We will reconstitute cellular and molecular network conferring oocyte dormancy using in vitro culture system. With a high resolution live-imaging system, we will reveal the dynamics of the gene network, cellular environment, and metabolic state for the acquisition of the oocyte dormancy.
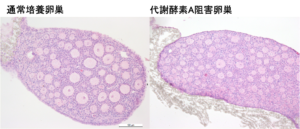
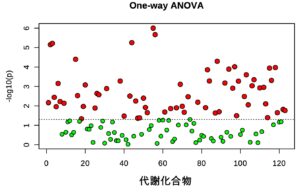
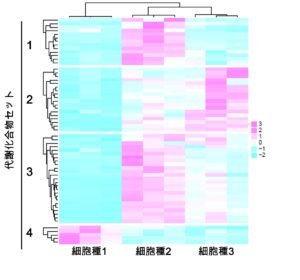
Using multi-omics analysis, including metabolomics and proteomics, we aim to elucidate changes in metabolic characteristics and their regulation throughout the reproductive lifespan of germ cells, and to modulate the reproductive lifespan via nutritional environments.
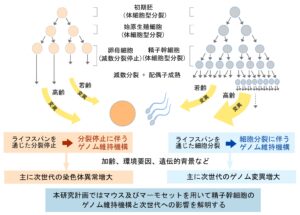
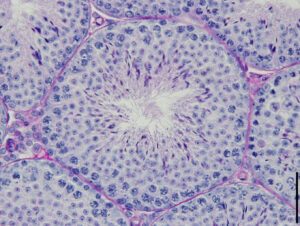
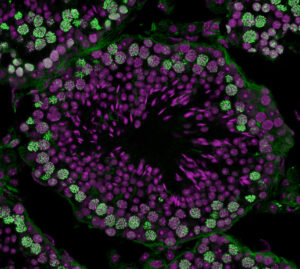
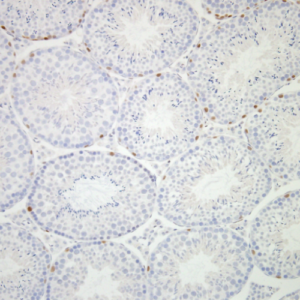
Focusing on the mechanisms of sperm stem cell maintenance and their population dynamics under homeostasis, this project is to elucidate the mechanisms by which stem cell clones harboring mutation harmful to the next generation expand in a non-neutral manner.
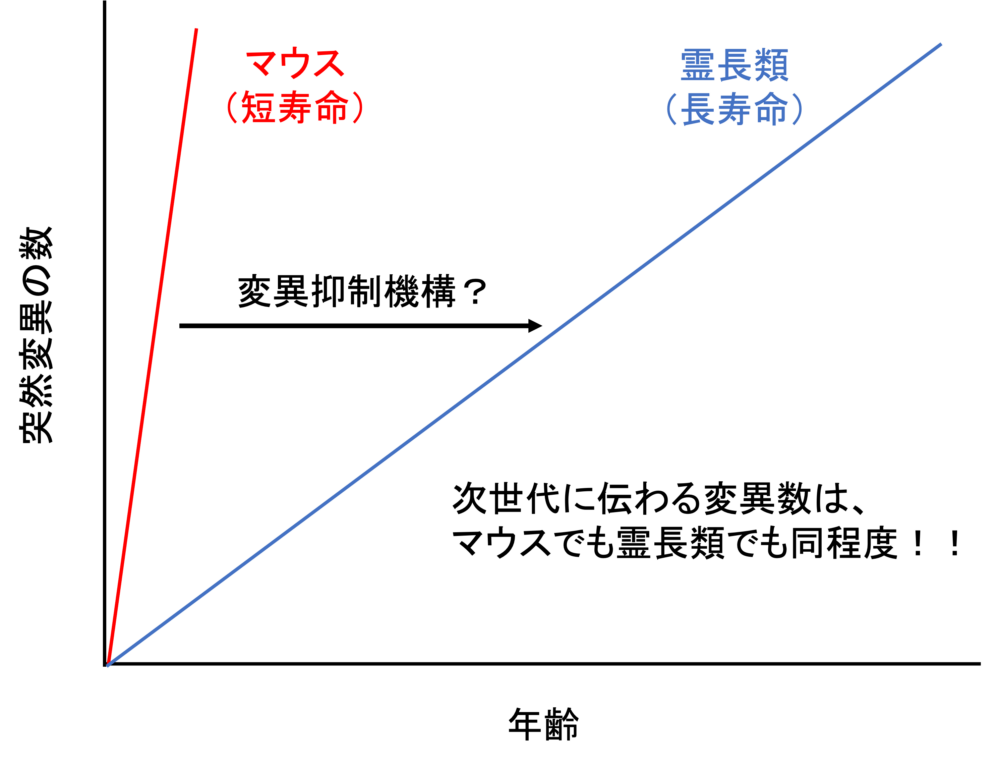
Our goal is to unravel the molecular basis of genome stability in spermatogonial stem cells and its functional implications for spermatogenesis and subsequent generations. Through a comparative analysis between mice and marmosets, we will investigate the regulatory dynamics and genome stability of primate spermatogonial stem cells.
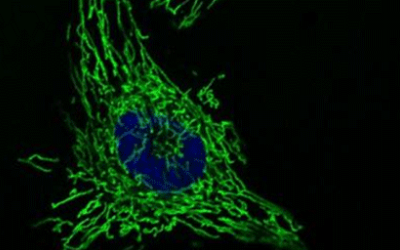
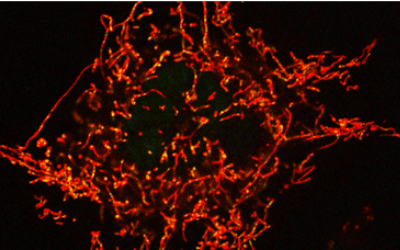
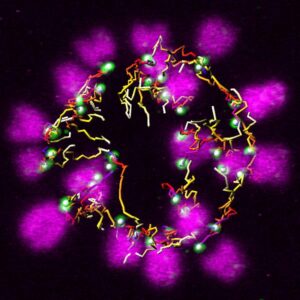
Mitochondrial DNAs (mtDNAs) are inherited from mother to child by maternal inheritance. We have observed an interesting phenomenon that the frequency of inheritance of mutant mtDNA to the next generation changes with aging of the mother mouse, and we will elucidate its mechanism.
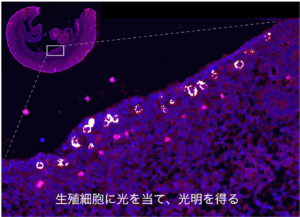
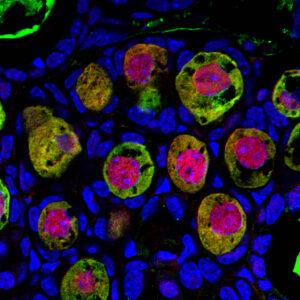
We will examine the spatiotemporal patterns governing germ cell function through the utilization of the Photo-Isolation Chemistry (PIC) technique, which enables the acquisition of local transcriptomic and epigenomic data from tissues and cells.
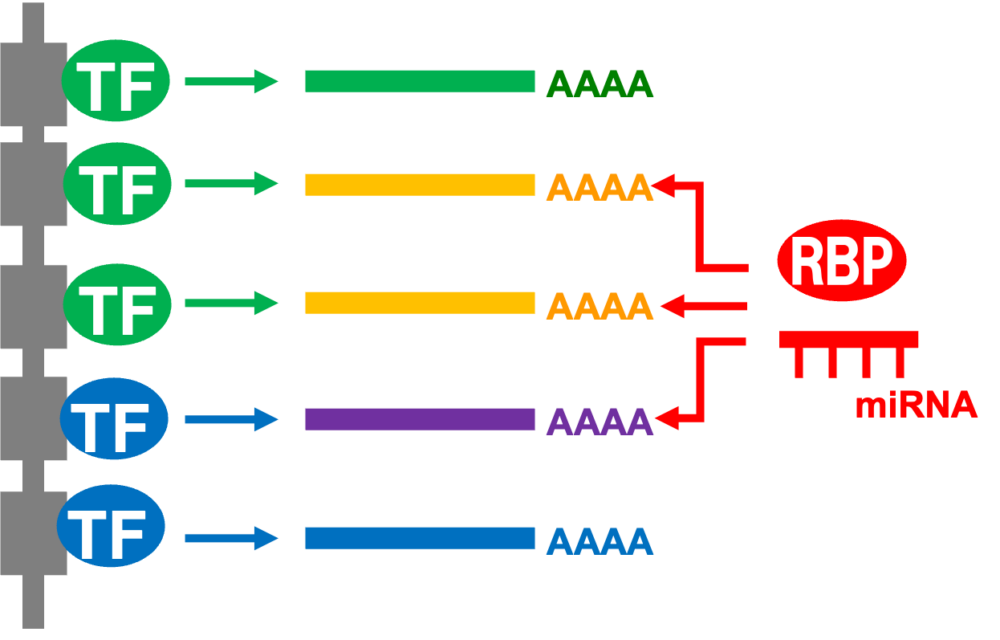
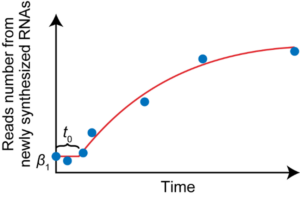
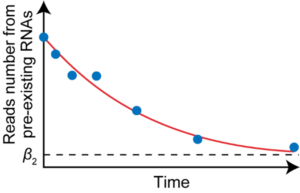
We will develop the RNA kinetics recording, which enables simultaneous quantification of the rates of RNA synthesis and degradation, for analysis at the single-cell level. Furthermore, by integrating this method with the PIC method, we will determine the spatially resolved rates of RNA synthesis and degradation in germ cells.
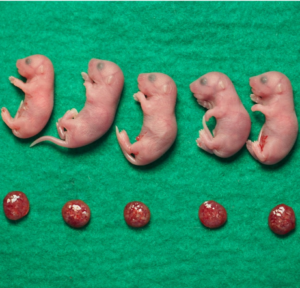
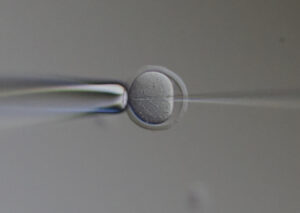
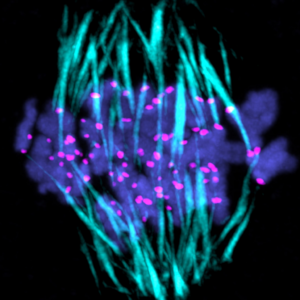
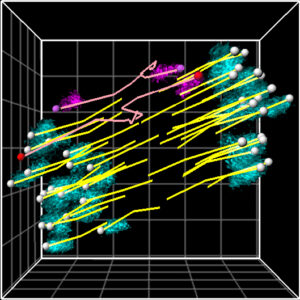
We will develop reproductive engineering technologies using nuclear transferred embryos as a model, and develop technologies to manipulate the reproductive lifespan, such as working with A01_01 on technologies to normalize chromosome segregation in aged oocytes.